Current Research Topics
Biofilms
In almost all natural habitats, microorganisms live in tightly associated communities called biofilms, rather than in the free-floating (planktonic) state. Natural biofilms are usually characterized by highly diverse species compositions. Biofilm cells are enclosed in a self-produced matrix of extracellular polymeric substances (EPS), protecting enclosed cells from harsh environmental conditions, such as desiccation, nutrient deficiency and disinfectants. Given their ubiquitous occurrence and their resistance to cleaning and disinfection procedures, biofilms are a relevant problem in food production. Biofilms on food-associated surfaces may lead to food contamination with pathogens or spoilage microorganisms, posing a threat to consumer health and food quality.
We aim to characterize the complex microbial composition of food-associated biofilms and investigate the dynamics of their formation. For this purpuse we use modern methods such as fluorescence in-situ hybridization (FISH).
Current example publication:
Weber, M., J. Liedtke, S. Plattes, A. Lipski. 2019. Bacterial community composition of biofilms in milking machines of two dairy farms assessed by a combination of culture-dependent and -independent methods. PLoS ONE 14(9):e0222238. Abstract


Cooperation between microorganisms
Due to their spatial proximity, biofilm cells interact with each other. This interaction can either be competitive, neutral, or cooperative. Cooperation involves the secretion of ‛public goods’ that confer benefit to the whole community. Division of labour when degrading complex molecules or utilization of metabolites of one another that would otherwise be toxic for the producer are other forms of cooperation. Biofilm inhabitants can communicate with each other by secreting signal molecules called ‛autoinducers’, whose local accumulation can alter the gene expression of the recipient. Such communication is termed quorum sensing (QS).
We aim to investigate mechanisms of cooperation between food-associated microorganisms using QS-reporter systems, analyses of growth and biofilm formation in mixed cultures as well as next generation sequencing.
Current example publication:
Hahne, J., A. Lipski. 2021. Growth interferences between bacterial strains from raw cow´s milk and their impact on growth of Listeria monocytogenes and Staphylococcus aureus. J. Appl. Microbiol. Abstract
Antibiotic resistances
The spread of antibiotic resistance genes in agricultural facilities is frequently associated with using these therapeutics in livestock breeding. The occurrence of resistances within these facilities and within animal-associated bacterial populations in comparison with those therapeutics actually used within these facilities reveals dependencies between the use of these therapeutics and the occurrence of resistances within livestock and plant associated bacterial communities. The analyses of parameters, which affect the spread of resistance genes in positive or negative are key for reducing or even preventing the transfer of resistance genes within bacterial communities in livestock breeding facilities.
Current example publication:
Weber, M., B. Göpfert, S. von Wezyk, M. Savin-Hoffmeyer, A. Lipski. 2023. Correlation between bacterial cell density and abundance of antibiotic resistance on milking machine surfaces assessed by cultivation and direct qPCR methods. Environ. Microbiol. Article
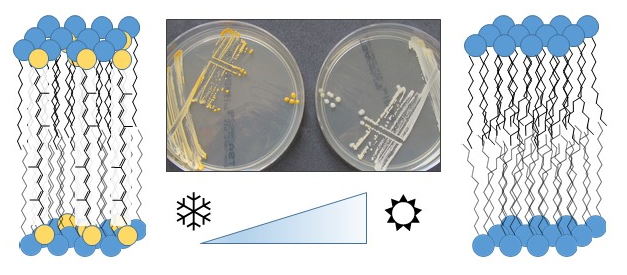
Adaptation of bacterial cell membranes to low temperatures
Growth at low-temperature conditions is challenging for microorganisms in various aspects. One of which is the regulation of cell membrane fluidity as an important requirement of fundamental biochemical activities such as energy production, signal processing, and transport. Besides the well-known adaptation of the membrane fatty acid profile, we analyze other membrane fluidization mechanisms for which only very little information is available so far. These mechanisms include the regulation of menaquinone and carotenoid concentration. Similar to the function of cholesterol these terpenoid-type lipids can affect membrane fluidity and membrane order and can ascertain the membrane functionality under low-temperature conditions.
Current example publication:
Flegler, A., A. Lipski, 2022. The C50 carotenoid bacterioruberin regulates membrane fluidity in pink-pigmented Arthrobacter species. Arch Microbiol 204, 70. Abstract
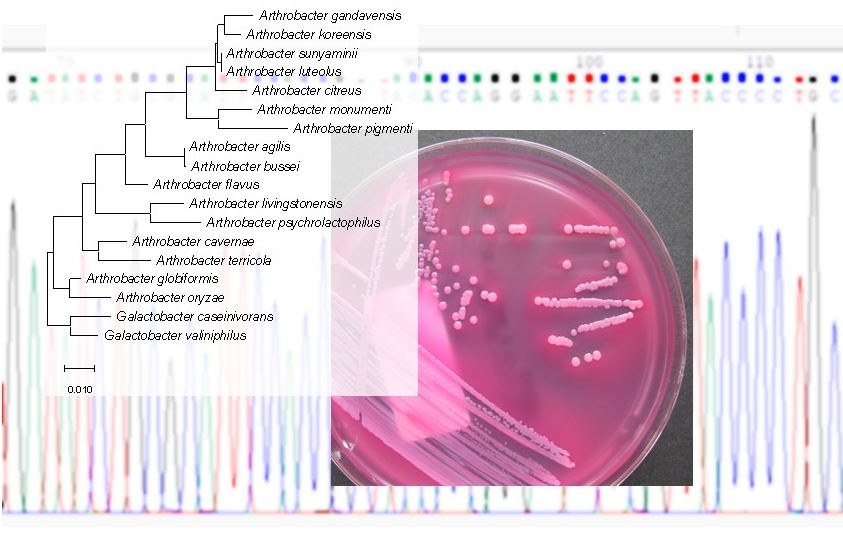
Taxonomy and systematics
In contrast to the eukaryotes, which diversity is well known, for prokaryotes several hundred of species are decribed per year and have to be integrated into the taxonomic system. Also in our projects we regularly find bacterial isolates which can not be assigned to any known bacterial species. These isolates are integrated into the taxonomic system based on the sequence of particular housekeeping genes, and characterized by phenotypic and genotypic properties. The phenotypic characterisation include analyzed of the lipid profile, including fatty acid profile, polar lipd pattern and quinone profile. In combination with a biochemical and morphological characterization and also based on genome sequences the characteriaztion let to the decription and proposal of new bacterial species. During the last 5 years our group was able to describe 11 new bacterial species or took part in the description.
Current example publication:
Heidler von Heilborn, D., L.-L. Nover, M. Weber, G. Höltzl, N. Gisch, C. Waldhans, M. Mittler, J. Kreyenschmidt, C. Woehle, B. Hüttel, A. Lipski. 2022. Polar lipid characterization and description of Chryseobacterium capnotolerans sp. nov., isolated from high CO2-containing atmosphere and emended descriptions of the genus Chryseobacterium, and the species C. balustinum, C. daecheongense, C. formosense, C. gleum, C. indologenes, C. joostei, C. scophthalmum and C. ureilyticum. Int. J. Syst. Evol. Microbiol. 72:005372. Abstract